Main Page: Difference between revisions
No edit summary |
No edit summary |
||
| Line 20: | Line 20: | ||
{{-}} | {{-}} | ||
== Management == | == Management == | ||
Revision as of 14:44, 20 July 2016
The Agrogenomics cluster is a High Performance Compute (HPC) infrastructure hosted by Wageningen University & Research Centre. It is open for use for all WUR research groups as well as other organizations, including companies, that have collaborative projects with WUR.
The Agrogenomics HPC was an initiative of the Breed4Food (B4F) consortium, consisting of the Animal Breeding and Genomics Centre (WU-Animal Breeding and Genomics and Wageningen Livestock Research) and four major breeding companies: Cobb-Vantress, CRV, Hendrix Genetics, and TOPIGS. Currently, in addition to the original partners, the HPC (HPC-Ag) is used by other groups from Wageningen UR (Bioinformatics, Centre for Crop Systems Analysis, Environmental Sciences Group, and Plant Research International) and plant breeding industry (Rijk Zwaan).
Rationale and Requirements for a new cluster

The Agrogenomics Cluster was originally conceived as being the 7th pillar of the Breed4Food programme. While the other six pillars revolve around specific research themes, the Cluster represents a joint infrastructure. The rationale behind the cluster is to enable the increasing computational needs in the field of genetics and genomics research, by creating a joint facility that will generate benefits of scale, thereby reducing cost. In addition, the joint infrastructure is intended to facilitate cross-organisational knowledge transfer. In that capacity, the HPC-Ag acts as a joint (virtual) laboratory where researchers - academic and applied - can benefit from each other's know-how. Lastly, the joint cluster, housed at Wageningen University campus, allows retaining vital and often confidential data sources in a controlled environment, something that cloud services such as Amazon Cloud or others usually can not guarantee.
Process of acquisition and financing

The Agrogenomics cluster was financed by CATAgroFood. The IT-Workgroup formulated a set of requirements that in the end were best met by an offer from Dell. ClusterVision was responsible for installing the cluster at the Theia server centre of FB-ICT.
Architecture of the cluster
Main Article: Architecture of the Agrogenomics HPC
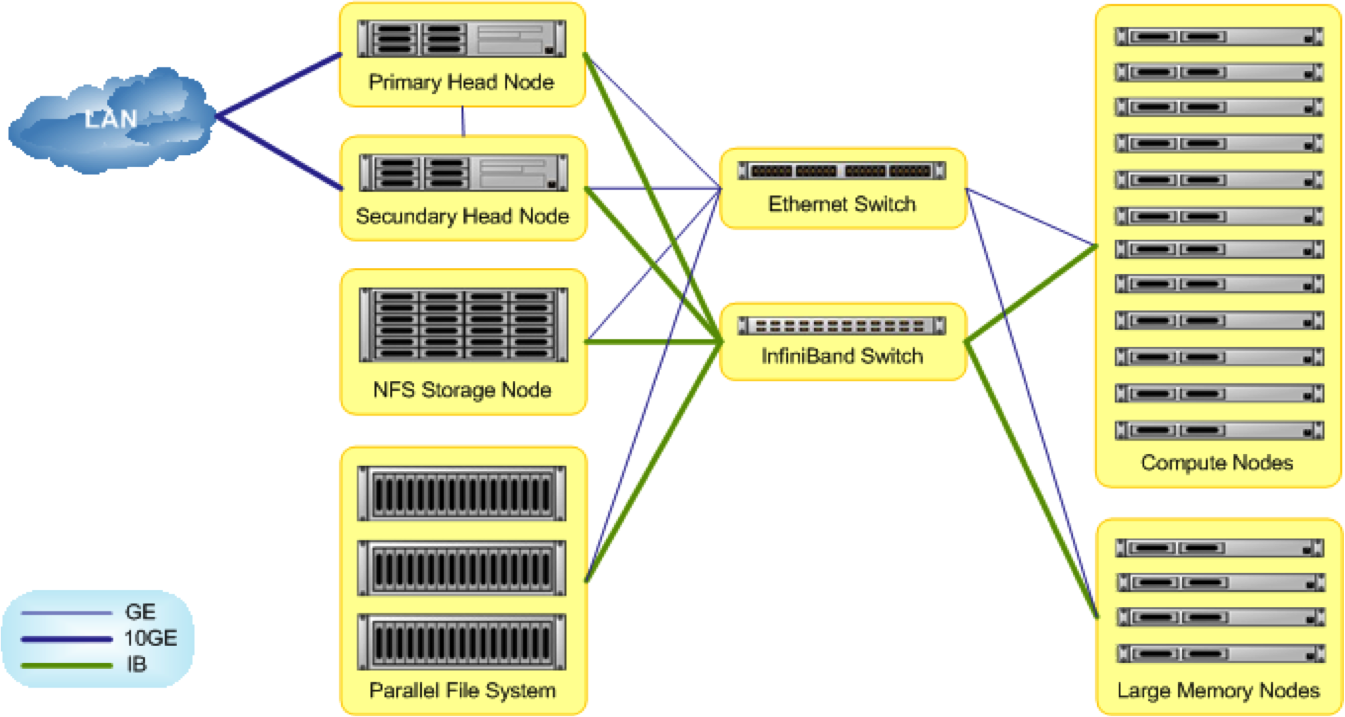
The new Agrogenomics HPC has a classic cluster architecture: state of the art Parallel File System (PSF), headnodes, compute nodes (of varying 'size'), all connected by superfast network connections (Infiniband). Implementation of the cluster will be done in stages. The initial stage includes a 600TB PFS, 48 slim nodes of 16 cores and 64GB RAM each, and 2 fat nodes of 64 cores and 1TB RAM each. The overall architecture, that include two head nodes in fall-over configuration and an infiniband network backbone, can be easily expanded by adding nodes and expanding the PFS. The cluster management software is designed to facilitate a heterogenous and evolving cluster.
Management
Project Leader of the HPC is Stephen Janssen (Wageningen UR,FB-IT, Service Management). Koen Pollmann (Wageningen UR,FB-IT, Infrastructure) and Gwen Dawes (Wageningen UR, FB-IT, Infrastructure) are responsible for Maintenance and Management.
Access Policy
Access needs to be granted actively (by creation of an account on the cluster by FB-IT). Use of resources is limited by the scheduler. Depending on availability of queues ('partitions') granted to a user, priority to the system's resources is regulated. Note that the use of the HPC-Ag is not free of charge. List price of CPU time and storage, and possible discounts on that list price for your organisation, can be retrieved from CAT-AGRO or FB-ICT.
Users
Using the HPC-Ag
Gaining access to the HPC-Ag
Access to the cluster and file transfer are done by ssh-based protocols.
Cluster Management Software and Scheduler
The HPC-Ag uses Bright Cluster Manager software for overall cluster management, and Slurm as job scheduler.
- Monitor cluster status with BCM
- Submit jobs with Slurm
- Be aware of how much work the cluster is under right now with 'node_usage_graph'
- Rosetta Stone of Workload Managers
Installation of software by users
- Installing domain specific software: installation by users
- Setting local variables
- Installing R packages locally
- Setting up and using a virtual environment for Python3
- Setting up and using a virtual environment for Python3.4 or higher
Installed software
Being in control of Environment parameters
- Using environment modules
- Setting local variables
- Set a custom temporary directory location
- Installing R packages locally
- Setting up and using a virtual environment for Python3
Controlling costs
Miscellaneous
- Bioinformatics tips, tricks, and workflows
- Convert between MediaWiki format and other formats
- GitLab: Create projects and add scripts
See also
- Maintenance and Management
- BCData
- Electronic mail discussion lists
- About ABGC
- High Performance Computing @ABGC
- Lustre Parallel File System layout