Population variant calling pipeline
Author: Carolina Pita Barros
Contact: carolina.pitabarros@wur.nl
ABG
For up-to-date documentation see here
Population level variant calling
Path to pipeline: /lustre/nobackup/WUR/ABGC/shared/PIPELINES/population-variant-calling
First follow the instructions here
Step by step guide on how to use my pipelines
Click here for an introduction to Snakemake
ABOUT
This is a pipeline that takes short reads aligned to a genome (in .bam format) and performs population level variant calling with Freebayes. It uses VEP to annotate the resulting VCF, calculates statistics, and calculates and plots a PCA.
It was developed to work with the results of this population mapping pipeline. There are a few Freebayes requirements that you need to take into account if you don't use the mapping pipeline mentioned above to map your reads. You should make sure that:
- Alignments have read groups
- Alignments are sorted
- Duplicates are marked
See here for more details.
Tools used
- Freebayes - variant calling using short reads
- bcftools - vcf statistics
- Plink - compute PCA
- R - Plot PCA
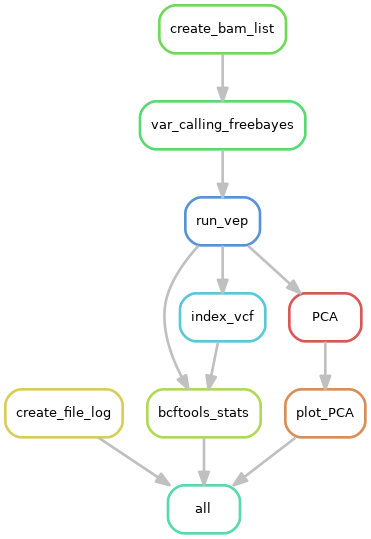
|
|---|
| Pipeline workflow |
Edit config.yaml with the paths to your files
<syntaxhighlight lang="yaml">ASSEMBLY: /path/to/fasta MAPPING_DIR: /path/to/bams/dir PREFIX: <prefix> OUTDIR: /path/to/outdir SPECIES: <species> NUM_CHRS: <number of chromosomes></syntaxhighlight>
- ASSEMBLY - path to genome fasta file
- MAPPING_DIR - path to directory with bam files to be used
- the pipeline will use all bam files in the directory, if you want to use a subset of those, create a file named
bam_list.txtthat contains the paths to the bam files you want to use. One path per line.
- the pipeline will use all bam files in the directory, if you want to use a subset of those, create a file named
<syntaxhighlight lang="text">/path/to/file.bam /path/to/file2.bam</syntaxhighlight>
- PREFIX - prefix for the created files
- OUTDIR - directory where snakemake will run and where the results will be written to
If you want the results to be written to this directory (not to a new directory), open config.yaml and comment out OUTDIR: /path/to/outdir
- SPECIES - species name to be used for VEP
- NUM_CHRS - number of chromosomes for your species (necessary for plink). ex: 38
ADDITIONAL SET UP
Configuring VEP
This pipeline uses VEP in offline mode, which increases performance. In order to use it in this mode, the cache for the species used needs to be installed:
For people using WUR's Anunna:
Check if the cache file for your species already exist in /lustre/nobackup/SHARED/cache/. If it doesn't, create it with
/usr/bin/perl /shared/apps/SHARED/ensembl-vep/INSTALL.pl --CACHEDIR /lustre/nobackup/SHARED/cache/ --AUTO c -n --SPECIES <species>
When multiple assemblies are found you need to run it again with --ASSEMBLY <assembly name>, where "assembly name" is the name of the assembly you want to use.
For those not from WUR:
You can install VEP with
conda install -c bioconda ensembl-vep
and install the cache with
vep_install --CACHEDIR <where/to/install/cache> --AUTO c -n --SPECIES <species>
When multiple assemblies are found you need to run it again with --ASSEMBLY <assembly name>, where "assembly name" is the name of the assembly you want to use.
In the Snakefile, in rule run_vep, replace /shared/apps/SHARED/ensembl-vep/vep with vep
Installing R packages
First load R: module load R/3.6.2
Enter the R environment by writing R and clicking enter. Install the packages:
list.of.packages <- c("optparse", "data.table", "ggplot2")
new.packages <- list.of.packages[!(list.of.packages %in% installed.packages()[,"Package"])]
if(length(new.packages)) install.packages(new.packages)
If you get an error like this:
Warning in install.packages(new.packages) : 'lib = "/shared/apps/R/3.6.2/lib64/R/library"' is not writable
Follow the instructions on how to install R packages locally here and try to install the packages again.
RESULTS
The most important files and directories are:
- <run_date>_files.txt dated file with an overview of the files used to run the pipeline (for documentation purposes)
- results directory that contains
- final_VCF directory with variant calling VCF files, as well as VCF stats
- {prefix}.vep.vcf.gz - final VCF file
- {prefix}.vep.vcf.gz.stats
- PCA PCA results and plot
- {prefix}.eigenvec and {prefix}.eigenval - file with PCA eigenvectors and eigenvalues, respectively
- {prefix}.pdf - PCA plot
- final_VCF directory with variant calling VCF files, as well as VCF stats
The VCF file has been filtered for QUAL > 20. Freebayes is ran with parameters --use-best-n-alleles 4 --min-base-quality 10 --min-alternate-fraction 0.2 --haplotype-length 0 --ploidy 2 --min-alternate-count 2. These parameters can be changed in the Snakefile.