Population structural variant calling pipeline: Difference between revisions
m added new config parameter - num of chrs |
m added info about split read support option |
||
| Line 24: | Line 24: | ||
* R - plot PCA | * R - plot PCA | ||
* SURVIVOR - basic SV stats | * SURVIVOR - basic SV stats | ||
* Python - add depth to vcf and create final table | * Python | ||
** | ** PyVcf - add depth to vcf and create final table | ||
** bamgroupreads.py + samblaster - create bam files with split and discordant reads | |||
{| | {| | ||
| Line 42: | Line 43: | ||
SPECIES: <species_name> | SPECIES: <species_name> | ||
PREFIX: <output name> | PREFIX: <output name> | ||
NUM_CHRS: <number of chromosomes></pre> | NUM_CHRS: <number of chromosomes> | ||
BWA_MEM_M: <Y/N></pre> | |||
* OUTDIR - directory where snakemake will run and where the results will be written to | * OUTDIR - directory where snakemake will run and where the results will be written to | ||
* READS_DIR - path to the directory that contains the reads | * READS_DIR - path to the directory that contains the reads | ||
| Line 58: | Line 61: | ||
* PREFIX - prefix for the created files | * PREFIX - prefix for the created files | ||
* NUM_CHRS - number of chromosomes for your species (necessary for plink). ex: 38 | * NUM_CHRS - number of chromosomes for your species (necessary for plink). ex: 38 | ||
* BWA_MEM_M - if you mapped your reads with `bwa mem` using the `-M` parameter and you want split read support in your VCF you need to run an extra step. For this write `Y`. | |||
For a more detailed explanation see here [https://carolinapb.github.io/2021-10-28-smoove-SR-support/ Smoove SR support] | |||
If you want the results to be written to this directory (not to a new directory), comment out or remove | If you want the results to be written to this directory (not to a new directory), comment out or remove | ||
Revision as of 09:50, 28 October 2021
Author: Carolina Pita Barros
Contact: carolina.pitabarros@wur.nl
ABG
First follow the instructions here:
Step by step guide on how to use my pipelines
Click here for an introduction to Snakemake
ABOUT
This is a pipeline to perform structural variant calling in a population using Smoove. It also runs VEP and performs PCA. In addition to the VCF with the SVs, you also get a .tsv file with some summarized information on the SVs: it includes allele frequency per population, as well as VEP annotation and depth fold change as described in duphold:
> DHBFC: fold-change for the variant depth relative to bins in the genome with similar GC-content.
> DHFFC: fold-change for the variant depth relative to Flanking regions.
Tools used:
- Smoove - SV calling
- VEP - determines the effect of the variants
- Plink - perform PCA
- R - plot PCA
- SURVIVOR - basic SV stats
- Python
- PyVcf - add depth to vcf and create final table
- bamgroupreads.py + samblaster - create bam files with split and discordant reads
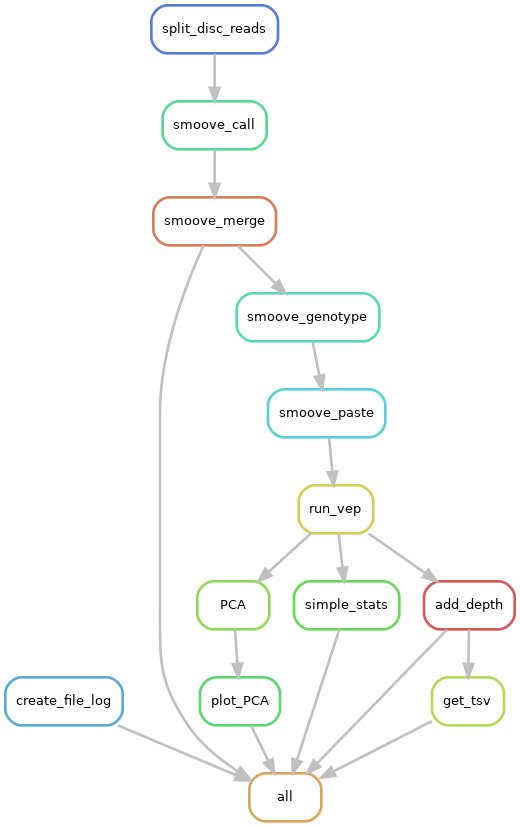
|
|---|
| Pipeline workflow |
Edit config.yaml with the paths to your files
OUTDIR: /path/to/output READS_DIR: /path/to/reads/ # don't add the reads files, just the directory where they are SAMPLE_LIST: /path/to/file REFERENCE: /path/to/assembly CONTIGS_IGNORE: /path/to/file SPECIES: <species_name> PREFIX: <output name> NUM_CHRS: <number of chromosomes> BWA_MEM_M: <Y/N>
- OUTDIR - directory where snakemake will run and where the results will be written to
- READS_DIR - path to the directory that contains the reads
- SAMPLE_LIST - three column csv with the sample name, name of the bam files to use in the second column and the name of the corresponding population on the third column. These bams should all be in the same directory (READS_DIR)
- Example:
sample1,sample1.bam,Pop1
sample2,sample2.bam,Pop1
sample3,sample3.bam,Pop2
sample4,sample4.bam,Pop2
Tip: use the name of the bam file without the .bam extension as the sample name. Ex: from sample1.bam to sample1
- REFERENCE - path to the assembly file
- CONTIGS_IGNORE - contigs to be excluded from SV calling (usually the small contigs)
- If you don't want to exclude contigs you'll need to edit the Snakefile to remove this line
--excludechroms {params.contigs} \
- If you don't want to exclude contigs you'll need to edit the Snakefile to remove this line
- SPECIES - species name to be used for VEP
- PREFIX - prefix for the created files
- NUM_CHRS - number of chromosomes for your species (necessary for plink). ex: 38
- BWA_MEM_M - if you mapped your reads with `bwa mem` using the `-M` parameter and you want split read support in your VCF you need to run an extra step. For this write `Y`.
For a more detailed explanation see here Smoove SR support
If you want the results to be written to this directory (not to a new directory), comment out or remove
OUTDIR: /path/to/outdir
ADDITIONAL SET UP
Configuring VEP
This pipeline uses VEP in offline mode, which increases performance. In order to use it in this mode, the cache for the species used needs to be installed:
Check if the cache file for your species already exist in /lustre/nobackup/SHARED/cache/. If it doesn’t, create it with
/usr/bin/perl /cm/shared/apps/SHARED/ensembl-vep/INSTALL.pl --CACHEDIR /lustre/nobackup/SHARED/cache/ --AUTO c -n --SPECIES <species>
When multiple assemblies are found you need to run it again with --ASSEMBLY <assembly name>, where “assembly name” is the name of the assembly you want to use.
Other option:
You can install VEP with
conda install -c bioconda ensembl-vep
and install the cache with
vep_install --CACHEDIR <where/to/install/cache> --AUTO c -n --SPECIES <species>
When multiple assemblies are found you need to run it again with --ASSEMBLY <assembly name>, where “assembly name” is the name of the assembly you want to use.
In the Snakefile, in rule run_vep, replace /cm/shared/apps/SHARED/ensembl-vep/vep with vep
Installing R packages
First load R:
module load R/3.6.2
Enter the R environment by writing
R
and clicking enter.
Install the packages:
list.of.packages <- c("optparse", "data.table", "ggplot2")
new.packages <- list.of.packages[!(list.of.packages %in% installed.packages()[,"Package"])]
if(length(new.packages)) install.packages(new.packages)
If you get an error like this:
Warning in install.packages(new.packages) : 'lib = "/cm/shared/apps/R/3.6.2/lib64/R/library"' is not writable
Follow the instructions on how to install R packages locally here and try to install the packages again.
RESULTS
- <run_date>_files.txt Dated file with an overview of the files used to run the pipeline (for documentation purposes)
- 2_merged
- {prefix}.smoove-counts.html - shows a summary of the number of reads before and after filtering
- 5_postprocessing directory that contains the final VCF file containing the structural variants found. This file has been annotated with VEP
- {prefix}.smoove.square.vep.vcf.gz - Final VCF - with VEP annotation, not filtered for quality
- {prefix}.smoove.square.vep.vcf.gz_summary.html - statistics from VEP
- {prefix}.nosex, {prefix}.log, {prefix}.eigenvec, {prefix}.eigenval - output files from the PCA
- {prefix}_DUP_DEL_INV_table.tsv - table with the most important information extracted from the VCF. Contains information about the SV, allele frequency for each population, VEP annotation and depth information. The variants have been filtered with Minimum Quality score = 30
- {prefix}_DUP_DEL_INV.vcf - vcf file with annotated duplications, deletions and inversions. It has been filtered with Minimum Quality score = 30 and the DEPTH* field was added
- {prefix}_BND.vcf - vcf file with variants annotated with BND
- 6_metrics directory that contains general stats about the number of SVs found
- FIGURES directory that contains the PCA plot
What you do with the results from this structural variant calling pipeline depends on your research question: a possible next step would be to explore the {prefix}_DUP_DEL_INV_table.tsv file and look at the largest SVs found (sort by SVLEN) or at a specific effect in the ANNOTATION column, such as “frameshift_variant”.
See VEP effect descriptions for a short description of the effects annotated by VEP.
- The DEPTH field in the vcf has six fields, corresponding to the average depth across all samples.
DEPTH=(DHBFC_1/1, DHBFC_0/1, DHBFC_0/0, DHFFC_1/1, DHFFC_0/1, DHFFC_0/0)
Depth fold change as described in duphold:
DHBFC: fold-change for the variant depth relative to bins in the genome with similar GC-content.
DHFFC: fold-change for the variant depth relative to Flanking regions.
These fields are also in the {prefix}_DUP_DEL_INV_table.tsv file