Installing R packages locally: Difference between revisions
Jump to navigation
Jump to search
m <source> to <pre> to fix code blocks |
No edit summary |
||
| (2 intermediate revisions by the same user not shown) | |||
| Line 30: | Line 30: | ||
install.packages("name-of-your-package",lib="~/R/library") | install.packages("name-of-your-package",lib="~/R/library") | ||
</pre> | </pre> | ||
== Installing and using a local library from a JuPyteR notebook == | |||
Click on the image. | |||
[[File:R_libpaths.png|thumb]] | |||
== See also == | == See also == | ||
* [[Control_R_environment_using_modules | Control R environment on the cluster using modules]] | * [[Control_R_environment_using_modules | Control R environment on the cluster using modules]] | ||
Latest revision as of 14:49, 9 April 2024
Specifying a local library search location
Specify a local library search location.
You can use several library trees of add-on packages. The easiest way to tell R to use these via a 'dotfile' by creating the following file '$HOME/.Renviron' (watch the quotes and ~ character):
R_LIBS_USER="~/R/library"
This specifies a keyword (R_LIBS_USER) which points to a colon-separated list of directories at which R library trees are rooted. You do not have to specify the default tree for R packages.
If necessary, create a place for your R libraries
mkdir ~/R ~/R/library # Only need do this once
Set your R library path
echo 'R_LIBS_USER="~/R/library"' > $HOME/.Renviron
Installing to a local library search location
Start up R:
R # Invoke R
Then, from the R environment, install the packages you require while pointing at the root R-package directory of choice. This example will install from CRAN.
install.packages("name-of-your-package",lib="~/R/library")
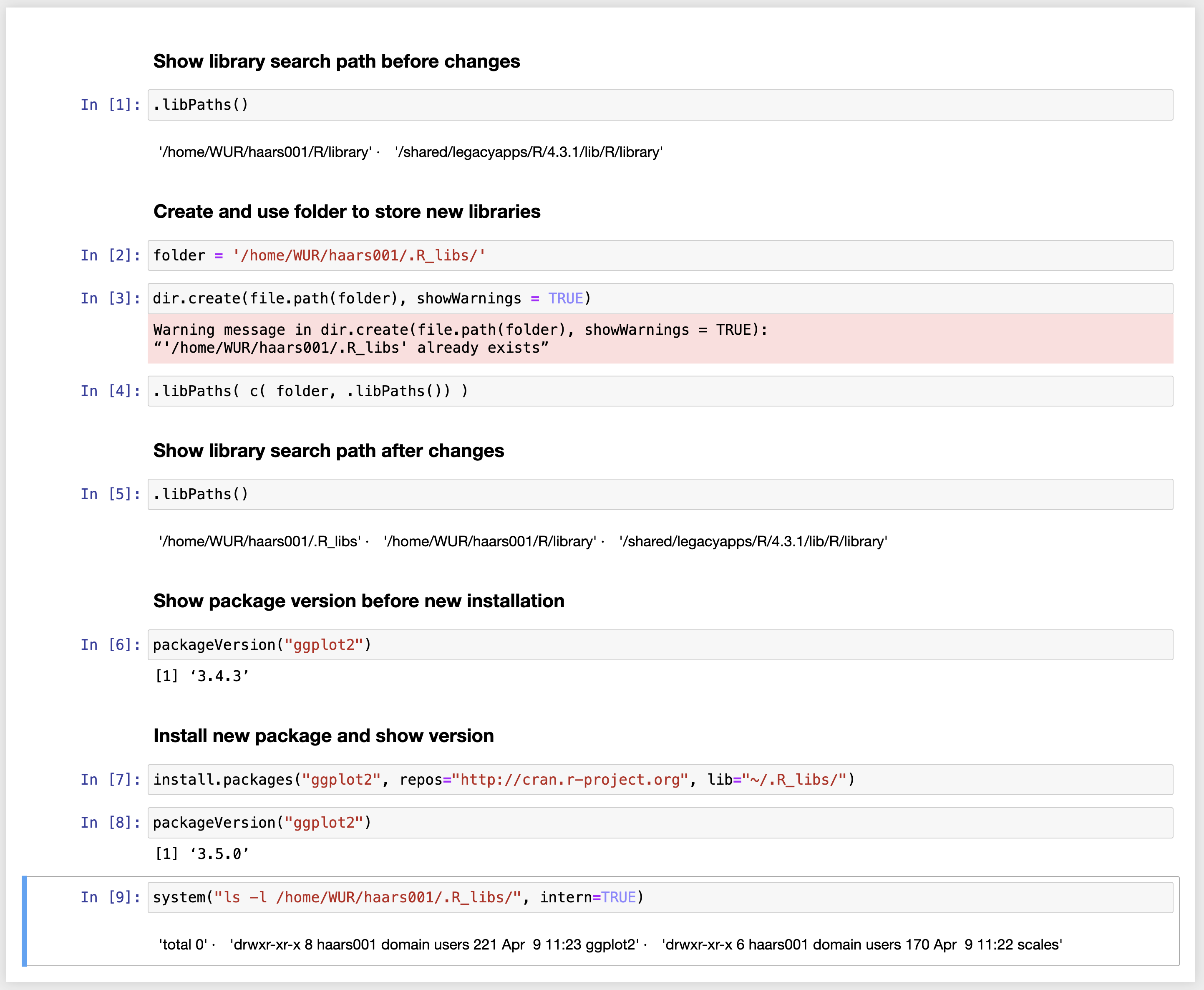
Installing and using a local library from a JuPyteR notebook
Click on the image.
See also
- Control R environment on the cluster using modules
- Using parallel R workloads under SLURM
- Bioinformatics tips, tricks, and workflows