Population mapping pipeline: Difference between revisions
Jump to navigation
Jump to search
added info about pipeline |
m added workflow image |
||
| Line 21: | Line 21: | ||
{| | {| | ||
!align="center"| [[File: | !align="center"| [[File:Population-mapping-workflow.png]] | ||
|- | |- | ||
|align="center"| ''Pipeline workflow'' | |align="center"| ''Pipeline workflow'' | ||
| Line 63: | Line 63: | ||
== RESULTS == | == RESULTS == | ||
* | * '''<run_date>_files.txt''' dated file with an overview of the files used to run the pipeline (for documentation purposes) | ||
* '''processed_reads''' directory with the bam files with the mapped reads for every sample | * '''processed_reads''' directory with the bam files with the mapped reads for every sample | ||
* '''mapping_stats''' directory containing the qualimap results and a summary of the qualimap results for all samples in <code>sample_quality_summary.tsv</code> | * '''mapping_stats''' directory containing the qualimap results and a summary of the qualimap results for all samples in <code>sample_quality_summary.tsv</code> | ||
** '''qualimap''' contains qualimap results per sample | ** '''qualimap''' contains qualimap results per sample | ||
Latest revision as of 13:55, 29 September 2021
Author: Carolina Pita Barros
Contact: carolina.pitabarros@wur.nl
ABG
First follow the instructions here:
Step by step guide on how to use my pipelines
Click here for an introduction to Snakemake
ABOUT
This is a pipeline to map short reads from several individuals to a reference assembly. It outputs the mapped reads and a qualimap report.
Tools used:
- Bwa - mapping
- Samtools - processing
- Qualimap - mapping summary
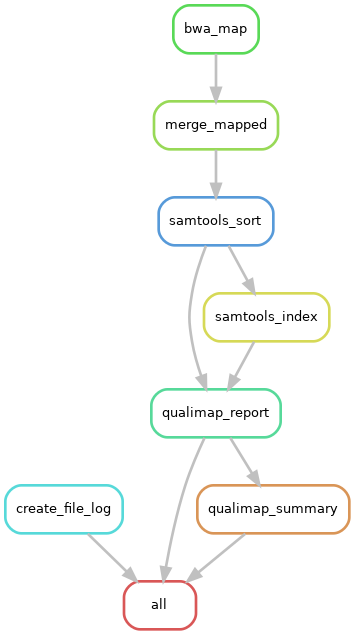
|
|---|
| Pipeline workflow |
Edit config.yaml with the paths to your files
ASSEMBLY: /path/to/assembly OUTDIR: /path/to/outdir PATHS_WITH_FILES: path1: /path/to/dir
- ASSEMBLY - path to the assembly file
- OUTDIR - directory where snakemake will run and where the results will be written to.
If you want the results to be written to this directory (not to a new directory), comment out OUTDIR: /path/to/outdir
- PATHS_WITH_FILES - directory that can contain subdirectories where the fq.gz reads are located. You can add several paths by adding
path2: /path/to/dirunderPATHS_WITH_FILES. (The line you add has to have indentation)
The script goes through the subdirectories of the directory you choose under PATHS_WITH_FILES looking for files with fq.gz extension.
Example: if path1: /lustre/nobackup/WUR/ABGC/shared/Chicken/Africa/X201SC20031230-Z01-F006_multipath, the subdirectory structure could be:
/lustre/nobackup/WUR/ABGC/shared/Chicken/Africa/X201SC20031230-Z01-F006_multipath
├── X201SC20031230-Z01-F006_1
│ └── raw_data
│ ├── a109_26_15_1_H
│ │ ├── a109_26_15_1_H_FDSW202597655-1r_HWFFFDSXY_L3_1.fq.gz
│ │ ├── a109_26_15_1_H_FDSW202597655-1r_HWFFFDSXY_L3_2.fq.gz
│ │ └── MD5.txt
│ └── a20_10_16_1_H
│ ├── a20_10_16_1_H_FDSW202597566-1r_HWFFFDSXY_L3_1.fq.gz
│ ├── a20_10_16_1_H_FDSW202597566-1r_HWFFFDSXY_L3_2.fq.gz
│ └── MD5.txt
└── X201SC20031230-Z01-F006_2
└── raw_data
├── a349_Be_17_1_C
│ ├── a349_Be_17_1_C_FDSW202597895-1r_HWFFFDSXY_L3_1.fq.gz
│ ├── a349_Be_17_1_C_FDSW202597895-1r_HWFFFDSXY_L3_2.fq.gz
│ └── MD5.txt
└── a360_Be_05_1_H
├── a360_Be_05_1_H_FDSW202597906-1r_HWFFFDSXY_L3_1.fq.gz
├── a360_Be_05_1_H_FDSW202597906-1r_HWFFFDSXY_L3_2.fq.gz
└── MD5.txt
RESULTS
- <run_date>_files.txt dated file with an overview of the files used to run the pipeline (for documentation purposes)
- processed_reads directory with the bam files with the mapped reads for every sample
- mapping_stats directory containing the qualimap results and a summary of the qualimap results for all samples in
sample_quality_summary.tsv- qualimap contains qualimap results per sample