Mapping and variant calling pipeline: Difference between revisions
No edit summary |
m info on how to concatenate fq files |
||
| (3 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
Author: Carolina Pita Barros | Author: Carolina Pita Barros <br /> | ||
Contact: carolina.pitabarros@wur.nl | Contact: carolina.pitabarros@wur.nl <br /> | ||
ABG | ABG<br /> | ||
Path to pipeline: /lustre/nobackup/WUR/ABGC/shared/PIPELINES/mapping-variant-calling | |||
[https://github.com/CarolinaPB/WUR_mapping-variant-calling Link to the repository] | [https://github.com/CarolinaPB/WUR_mapping-variant-calling Link to the repository] | ||
| Line 16: | Line 18: | ||
==== Tools used: ==== | ==== Tools used: ==== | ||
* Bwa - mapping | * Bwa-mem2 - mapping | ||
* Samtools - processing | * Samtools - processing | ||
* Qualimap - mapping summary | * Qualimap - mapping summary | ||
* Freebayes - variant calling | * Freebayes - variant calling | ||
* Bcftools - VCF statistics | |||
{| | |||
!align="center"| [[File:mapping-variant-calling-workflow.png]] | |||
|- | |||
|align="center"| ''Pipeline workflow'' | |||
|} | |||
=== Edit config.yaml with the paths to your files === | === Edit config.yaml with the paths to your files === | ||
| Line 32: | Line 41: | ||
* PREFIX - prefix for the final mapped reads file | * PREFIX - prefix for the final mapped reads file | ||
If you want the results to be written to this directory (not to a new directory), | If you want the results to be written to this directory (not to a new directory), comment out <pre>OUTDIR: /path/to/output</pre> | ||
For the mapping step you should have one _1 fastq file and one _2 fastq file in <code>READS_DIR</code>. If you have several _1 and _2 fastq files from the same sample, you can combine them so you have one file for all _1 reads and one for all the _2 reads. This can be done by concatenating them using <code>cat</code>, if the original files are not compressed (<code>fastq</code> or <code>fq</code> extension), or <code>zcat</code> if the original files are compressed (<code>fastq.gz</code> or <code>fq.gz</code> extension). Example where your files are in the same directory and are compressed: | |||
<pre>zcat *_1.fastq.gz > <new file name>_1.fastq.gz | |||
zcat *_2.fastq.gz > <new file name>_2.fastq.gz</pre> | |||
== RESULTS == | == RESULTS == | ||
| Line 40: | Line 53: | ||
* '''sorted_reads''' directory with the file containing the mapped reads | * '''sorted_reads''' directory with the file containing the mapped reads | ||
* '''results''' directory containing the qualimap results | * '''results''' directory containing the qualimap results | ||
* '''variant_calling''' directory containing the variant calling VCF file | * '''variant_calling''' directory containing the variant calling VCF file and a file with with VCF statistics | ||
Latest revision as of 12:56, 22 November 2021
Author: Carolina Pita Barros
Contact: carolina.pitabarros@wur.nl
ABG
Path to pipeline: /lustre/nobackup/WUR/ABGC/shared/PIPELINES/mapping-variant-calling
First follow the instructions here:
Step by step guide on how to use my pipelines
Click here for an introduction to Snakemake
ABOUT
This is a pipeline to map short reads to a reference assembly. It outputs the mapped reads, a qualimap report and does variant calling.
Tools used:
- Bwa-mem2 - mapping
- Samtools - processing
- Qualimap - mapping summary
- Freebayes - variant calling
- Bcftools - VCF statistics
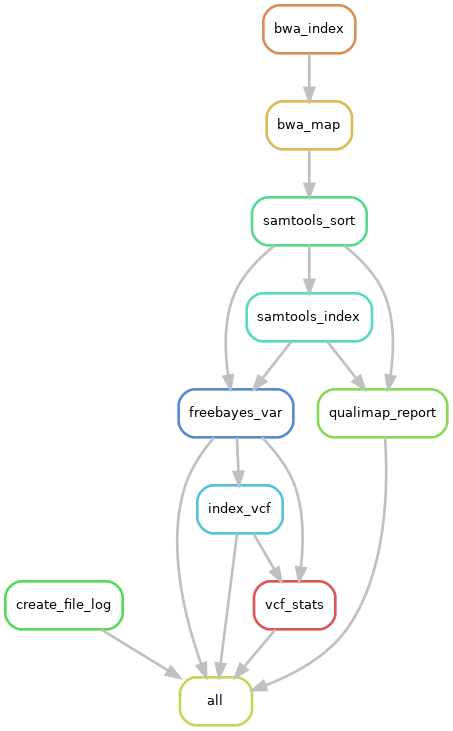
|
|---|
| Pipeline workflow |
Edit config.yaml with the paths to your files
OUTDIR: /path/to/output READS_DIR: /path/to/reads/ # don't add the reads files, just the directory where they are ASSEMBLY: /path/to/assembly PREFIX: <output name>
- OUTDIR - directory where snakemake will run and where the results will be written to
- READS_DIR - path to the directory that contains the reads
- ASSEMBLY - path to the assembly file
- PREFIX - prefix for the final mapped reads file
If you want the results to be written to this directory (not to a new directory), comment out
OUTDIR: /path/to/output
For the mapping step you should have one _1 fastq file and one _2 fastq file in READS_DIR. If you have several _1 and _2 fastq files from the same sample, you can combine them so you have one file for all _1 reads and one for all the _2 reads. This can be done by concatenating them using cat, if the original files are not compressed (fastq or fq extension), or zcat if the original files are compressed (fastq.gz or fq.gz extension). Example where your files are in the same directory and are compressed:
zcat *_1.fastq.gz > <new file name>_1.fastq.gz zcat *_2.fastq.gz > <new file name>_2.fastq.gz
RESULTS
- dated file with an overview of the files used to run the pipeline (for documentation purposes)
- sorted_reads directory with the file containing the mapped reads
- results directory containing the qualimap results
- variant_calling directory containing the variant calling VCF file and a file with with VCF statistics