Nanopore assembly and variant calling: Difference between revisions
added info about nanopore pipeline |
m added link to github repo |
||
| (2 intermediate revisions by the same user not shown) | |||
| Line 3: | Line 3: | ||
ABG<br /> | ABG<br /> | ||
For up-to-date documentation see [https://github.com/CarolinaPB/nanopore-assembly here] | |||
= Assemble nanopore reads and do variant calling with short and long reads = | = Assemble nanopore reads and do variant calling with short and long reads = | ||
Path to pipeline: /lustre/nobackup/WUR/ABGC/shared/PIPELINES/nanopore-assembly | Path to pipeline: /lustre/nobackup/WUR/ABGC/shared/PIPELINES/nanopore-assembly | ||
| Line 14: | Line 15: | ||
This is a pipeline that uses <code>Flye</code> to create a nanopore assembly. It also does variant calling with long and short reads.<br /> | This is a pipeline that uses <code>Flye</code> to create a nanopore assembly. It also does variant calling with long and short reads.<br /> | ||
The pipeline starts by using <code>porechop</code> to trim the adaptors, then it uses <code>Flye</code> to create the assembly. After that, <code>ntLink-arks</code> from <code>Lonstitch</code> is used to scaffold the assembly using the nanopore reads. The scaffolded assembly is polished with <code>polca</code>. <code> | The pipeline starts by using <code>porechop</code> to trim the adaptors, then it uses <code>Flye</code> to create the assembly. After that, <code>ntLink-arks</code> from <code>Lonstitch</code> is used to scaffold the assembly using the nanopore reads. The scaffolded assembly is polished with <code>polca</code>. <code>Bwa-mem2</code> is used to map the short reads to the assembly and <code>Freebayes</code> to do variant calling using these reads. <code>Minimap2</code> is used to map the long reads to the assembly, and <code>longshot</code> for variant calling using these. In the end, in addition to your assembly and variant calling results, you'll also get assembly statistics and busco scores before and after the polishing. | ||
In the end, in addition to your assembly and variant calling results, you'll also get assembly statistics and busco scores before and after the polishing. | |||
==== Tools used: ==== | ==== Tools used: ==== | ||
| Line 24: | Line 24: | ||
* [https://github.com/bcgsc/longstitch LongStitch (ntLink-arks)] - scaffolding with nanopore reads | * [https://github.com/bcgsc/longstitch LongStitch (ntLink-arks)] - scaffolding with nanopore reads | ||
* [https://busco.ezlab.org/ BUSCO] - assess assembly completeness | * [https://busco.ezlab.org/ BUSCO] - assess assembly completeness | ||
* [https://github.com/alekseyzimin/masurca MaSuRCA (polca)] - polish assembly | * [https://github.com/alekseyzimin/masurca MaSuRCA (polca)] - polish assembly | ||
* Python - get assembly stats | * Python - get assembly stats | ||
* [https://github.com/lh3/minimap2 Minimap2] - map long reads to reference. Genome alignment | * [https://github.com/lh3/minimap2 Minimap2] - map long reads to reference. Genome alignment | ||
* [http://www.htslib.org/ Samtools] - sort and index mapped reads and vcf files | * [http://www.htslib.org/ Samtools] - sort and index mapped reads and vcf files | ||
* [https://github.com/pjedge/longshot Longshot] - variant calling with nanopore reads | * [https://github.com/pjedge/longshot Longshot] - variant calling with nanopore reads | ||
* [https://github.com/bwa-mem2/bwa-mem2 Bwa-mem2] - map short reads to reference | |||
* [https://github.com/freebayes/freebayes Freebayes] - variant calling using short reads | |||
* [https://samtools.github.io/bcftools/bcftools.html bcftools] - vcf statistics | * [https://samtools.github.io/bcftools/bcftools.html bcftools] - vcf statistics | ||
* R - [https://github.com/tpoorten/dotPlotly pafCoordsDotPlotly] - plot genome alignment | * R - [https://github.com/tpoorten/dotPlotly pafCoordsDotPlotly] - plot genome alignment | ||
| Line 103: | Line 105: | ||
*** {prefix}_longreads.vcf.gz | *** {prefix}_longreads.vcf.gz | ||
*** {prefix}_longreads.vcf.gz.stats | *** {prefix}_longreads.vcf.gz.stats | ||
Both the short reads and long reads variant calling VCF is filtered for <code>QUAL > 20 </code>. | |||
Freebayes (short read var calling) is ran with parameters <code>--use-best-n-alleles 4 --min-base-quality 10 --min-alternate-fraction 0.2 --haplotype-length 0 --ploidy 2 --min-alternate-count 2 </code>. For more details check the Snakefile. | |||
** '''genome_alignment''' directory with results and figure from whole genome alignment | ** '''genome_alignment''' directory with results and figure from whole genome alignment | ||
*** {prefix}_{species}.png | *** {prefix}_{species}.png | ||
Latest revision as of 15:36, 13 June 2022
Author: Carolina Pita Barros
Contact: carolina.pitabarros@wur.nl
ABG
For up-to-date documentation see here
Assemble nanopore reads and do variant calling with short and long reads
Path to pipeline: /lustre/nobackup/WUR/ABGC/shared/PIPELINES/nanopore-assembly
First follow the instructions here:
Step by step guide on how to use my pipelines
Click here for an introduction to Snakemake
ABOUT
This is a pipeline that uses Flye to create a nanopore assembly. It also does variant calling with long and short reads.
The pipeline starts by using porechop to trim the adaptors, then it uses Flye to create the assembly. After that, ntLink-arks from Lonstitch is used to scaffold the assembly using the nanopore reads. The scaffolded assembly is polished with polca. Bwa-mem2 is used to map the short reads to the assembly and Freebayes to do variant calling using these reads. Minimap2 is used to map the long reads to the assembly, and longshot for variant calling using these. In the end, in addition to your assembly and variant calling results, you'll also get assembly statistics and busco scores before and after the polishing.
Tools used:
- Porechop - trim adaptors
- Flye - assembly
- Seqtk - convert fasta to one line fasta
- LongStitch (ntLink-arks) - scaffolding with nanopore reads
- BUSCO - assess assembly completeness
- MaSuRCA (polca) - polish assembly
- Python - get assembly stats
- Minimap2 - map long reads to reference. Genome alignment
- Samtools - sort and index mapped reads and vcf files
- Longshot - variant calling with nanopore reads
- Bwa-mem2 - map short reads to reference
- Freebayes - variant calling using short reads
- bcftools - vcf statistics
- R - pafCoordsDotPlotly - plot genome alignment
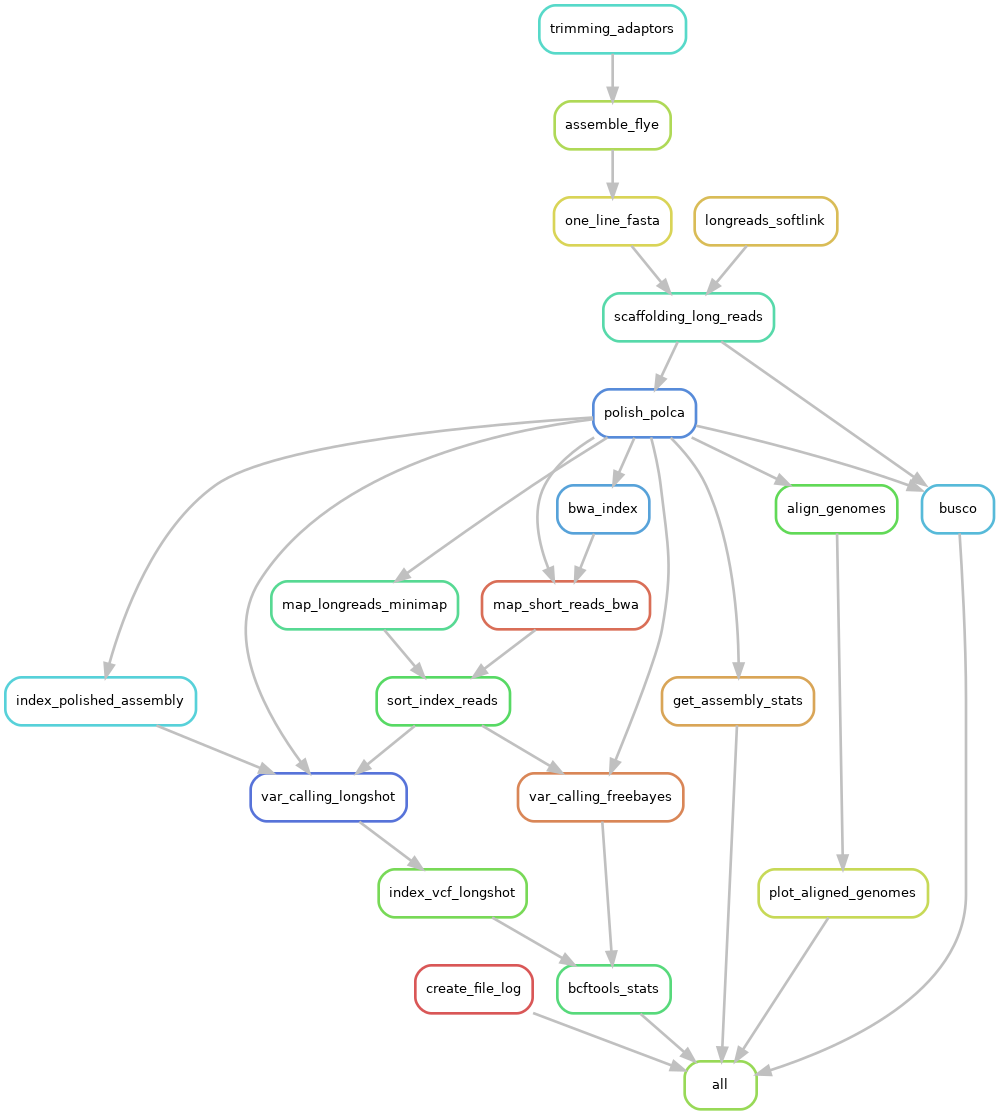
|
|---|
| Pipeline workflow |
Edit config.yaml with the paths to your files
LONGREADS: <nanopore_reads.fq.gz> SHORTREADS: - /path/to/short/reads_1.fq.gz - /path/to/short/reads_2.fq.gz GENOME_SIZE: <approximate genome size> PREFIX: <prefix> OUTDIR: /path/to/outdir BUSCO_LINEAGE: - <lineage> # genome alignment parameters: COMPARISON_GENOME: <species>: /path/to/genome/fasta # filter alignments less than cutoff X bp MIN_ALIGNMENT_LENGTH: 10000 MIN_QUERY_LENGTH: 50000
- LONGREADS - name of file with long reads. This file should be in the working directory (where this config and the Snakefile are)
- SHORTREADS - paths to short reads fq.gz
- GENOME_SIZE - approximate genome size
haploid genome size (bp)(e.g. '3e9' for human genome)from longstitch - PREFIX - prefix for the created files
- OUTDIR - directory where snakemake will run and where the results will be written to
If you want the results to be written to this directory (not to a new directory), open config.yaml and comment out OUTDIR: /path/to/outdir
- BUSCO_LINEAGE - lineage used for busco. Can be one or more (one per line). To see available lineages run
busco --list-datasets - COMPARISON_GENOME - genome for whole genome comparison. Add your species name and the path to the fasta file. ex:
chicken: /path/to/chicken.fna.gz. You can add several genomes, one on each line.- If you don't want to run the genome alignment step, comment out
COMPARISON_GENOME: <species>: /path/to/genome/fasta
- MIN_ALIGNMENT_LENGTH and MIN_QUERY_LENGTH - parameters for plotting. If your plot is coming out blank or if there's an error with the plotting step, try lowering these thresholds. This happens because the alignments are not large enough.
If you have your long reads in several fastq files and need to create one file compressed file with all the reads:
- In your pipeline directory create one file with all the reads
cat /path/to/fastq/directory/*.fastq > <name of file>.fq
- Compress the file you just created:
gzip <name of file>.fq
After installing the conda environment (first step of this guide) you'll need to edit the polca.sh file.
First go to the directory where miniconda3 is installed (usually your home directory). Go to /<home>/miniconda/envs/<env_name>/bin and open the file polca.sh. In my case the path looks like this: /home/WUR/<username>/miniconda3/envs/<env_name>/bin/. In your editor open polca.sh and replace this line:
$SAMTOOLS sort -m $MEM -@ $NUM_THREADS <(samtools view -uhS $BASM.unSorted.sam) $BASM.alignSorted 2>>samtools.err && \
With this:
$SAMTOOLS sort -m $MEM -@ $NUM_THREADS <(samtools view -uhS $BASM.unSorted.sam) -o $BASM.alignSorted.bam 2>>samtools.err && \
RESULTS
The most important files are and directories are:
- <run_date>_files.txt dated file with an overview of the files used to run the pipeline (for documentation purposes)
- results directory that contains
- {prefix}_oneline.k32.w100.ntLink-arks.longstitch-scaffolds.fa.PolcaCorrected.fa final assembly
- assembly_stats_<prefix>.txt file with assembly statistics for the final assembly
- variant_calling directory with variant calling VCF files with long and short reads, as well as VCF stats
- {prefix}_shortreads.vcf.gz
- {prefix}_shortreads.vcf.gz.stats
- {prefix}_longreads.vcf.gz
- {prefix}_longreads.vcf.gz.stats
Both the short reads and long reads variant calling VCF is filtered for QUAL > 20 .
Freebayes (short read var calling) is ran with parameters --use-best-n-alleles 4 --min-base-quality 10 --min-alternate-fraction 0.2 --haplotype-length 0 --ploidy 2 --min-alternate-count 2 . For more details check the Snakefile.
- genome_alignment directory with results and figure from whole genome alignment
- {prefix}_{species}.png
- genome_alignment directory with results and figure from whole genome alignment
- mapped directory that contains the bam file with long reads mapped to the new assembly
- {prefix}_longreads.mapped.sorted.bam
- busco_{prefix}_before_polish_ and busco_{prefix}_after_polish directories - contain busco results before and after polishing
- short_summary.specific.{lineage}.{prefix}_before_polish.txt
- short_summary.specific.{lineage}.{prefix}_after_polish.txt"
- other_files - directory containing other files created during the pipeline
- assembly - directory containing files created during the assembly step