Population variant calling pipeline: Difference between revisions
added info on pipeline |
m added workflow image |
||
| Line 31: | Line 31: | ||
{| | {| | ||
!align="center"| [[File: | !align="center"| [[File:population-var-calling-workflow.png]] | ||
|- | |- | ||
|align="center"| ''Pipeline workflow'' | |align="center"| ''Pipeline workflow'' | ||
Revision as of 14:45, 2 March 2022
Population level variant calling
Path to pipeline: /lustre/nobackup/WUR/ABGC/shared/PIPELINES/population-variant-calling
First follow the instructions here
Step by step guide on how to use my pipelines
Click here for an introduction to Snakemake
ABOUT
This is a pipeline that takes short reads aligned to a genome (in .bam format) and performs population level variant calling with Freebayes. It uses VEP to annotate the resulting VCF, calculates statistics, and calculates and plots a PCA.
It was developed to work with the results of this population mapping pipeline. There are a few Freebayes requirements that you need to take into account if you don't use the mapping pipeline mentioned above to map your reads. You should make sure that:
- Alignments have read groups
- Alignments are sorted
- Duplicates are marked
See here for more details.
Tools used
- Freebayes - variant calling using short reads
- bcftools - vcf statistics
- Plink - compute PCA
- R - Plot PCA
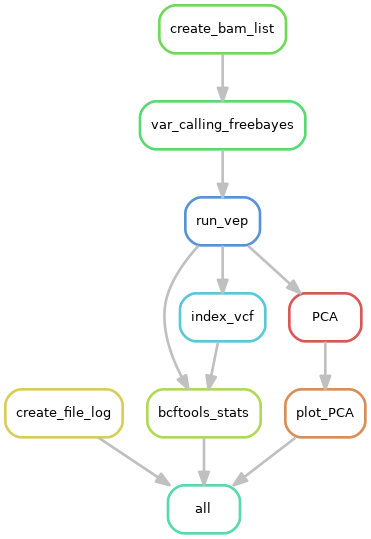
|
|---|
| Pipeline workflow |
Edit config.yaml with the paths to your files
<syntaxhighlight lang="yaml">ASSEMBLY: /path/to/fasta MAPPING_DIR: /path/to/bams/dir PREFIX: <prefix> OUTDIR: /path/to/outdir SPECIES: <species> NUM_CHRS: <number of chromosomes></syntaxhighlight>
- ASSEMBLY - path to genome fasta file
- MAPPING_DIR - path to directory with bam files to be used
- the pipeline will use all bam files in the directory, if you want to use a subset of those, create a file named
bam_list.txtthat contains the paths to the bam files you want to use. One path per line.
- the pipeline will use all bam files in the directory, if you want to use a subset of those, create a file named
<syntaxhighlight lang="text">/path/to/file.bam /path/to/file2.bam</syntaxhighlight>
- PREFIX - prefix for the created files
- OUTDIR - directory where snakemake will run and where the results will be written to
If you want the results to be written to this directory (not to a new directory), open config.yaml and comment out OUTDIR: /path/to/outdir
- SPECIES - species name to be used for VEP
- NUM_CHRS - number of chromosomes for your species (necessary for plink). ex: 38
RESULTS
The most important files and directories are:
- <run_date>_files.txt dated file with an overview of the files used to run the pipeline (for documentation purposes)
- results directory that contains
- final_VCF directory with variant calling VCF files, as well as VCF stats
- {prefix}.vep.vcf.gz - final VCF file
- {prefix}.vep.vcf.gz.stats
- PCA PCA results and plot
- {prefix}.eigenvec and {prefix}.eigenval - file with PCA eigenvectors and eigenvalues, respectively
- {prefix}.pdf - PCA plot
- final_VCF directory with variant calling VCF files, as well as VCF stats
The VCF file has been filtered for QUAL > 20. Freebayes is ran with parameters --use-best-n-alleles 4 --min-base-quality 10 --min-alternate-fraction 0.2 --haplotype-length 0 --ploidy 2 --min-alternate-count 2. These parameters can be changed in the Snakefile.