Mapping and variant calling pipeline: Difference between revisions
Jump to navigation
Jump to search
mNo edit summary |
m added pipeline workflow, small details |
||
| Line 20: | Line 20: | ||
* Qualimap - mapping summary | * Qualimap - mapping summary | ||
* Freebayes - variant calling | * Freebayes - variant calling | ||
* Bcftools - VCF statistics | |||
{| | |||
!align="center"| [[File:mapping-variant-calling-workflow.png]] | |||
|- | |||
|align="center"| ''Pipeline workflow'' | |||
|} | |||
=== Edit config.yaml with the paths to your files === | === Edit config.yaml with the paths to your files === | ||
| Line 32: | Line 39: | ||
* PREFIX - prefix for the final mapped reads file | * PREFIX - prefix for the final mapped reads file | ||
If you want the results to be written to this directory (not to a new directory), | If you want the results to be written to this directory (not to a new directory), comment out <pre>READS_DIR: /path/to/reads/</pre> | ||
<pre> | |||
== RESULTS == | == RESULTS == | ||
| Line 40: | Line 45: | ||
* '''sorted_reads''' directory with the file containing the mapped reads | * '''sorted_reads''' directory with the file containing the mapped reads | ||
* '''results''' directory containing the qualimap results | * '''results''' directory containing the qualimap results | ||
* '''variant_calling''' directory containing the variant calling VCF file | * '''variant_calling''' directory containing the variant calling VCF file and a file with with VCF statistics | ||
Revision as of 12:46, 22 October 2021
Author: Carolina Pita Barros
Contact: carolina.pitabarros@wur.nl
ABG
First follow the instructions here:
Step by step guide on how to use my pipelines
Click here for an introduction to Snakemake
ABOUT
This is a pipeline to map short reads to a reference assembly. It outputs the mapped reads, a qualimap report and does variant calling.
Tools used:
- Bwa - mapping
- Samtools - processing
- Qualimap - mapping summary
- Freebayes - variant calling
- Bcftools - VCF statistics
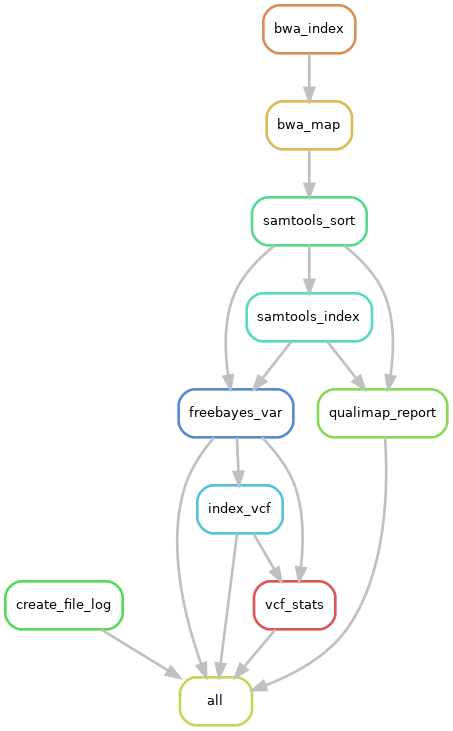
|
|---|
| Pipeline workflow |
Edit config.yaml with the paths to your files
OUTDIR: /path/to/output READS_DIR: /path/to/reads/ # don't add the reads files, just the directory where they are ASSEMBLY: /path/to/assembly PREFIX: <output name>
- OUTDIR - directory where snakemake will run and where the results will be written to
- READS_DIR - path to the directory that contains the reads
- ASSEMBLY - path to the assembly file
- PREFIX - prefix for the final mapped reads file
If you want the results to be written to this directory (not to a new directory), comment out
READS_DIR: /path/to/reads/
RESULTS
- dated file with an overview of the files used to run the pipeline (for documentation purposes)
- sorted_reads directory with the file containing the mapped reads
- results directory containing the qualimap results
- variant_calling directory containing the variant calling VCF file and a file with with VCF statistics